Python, R, Bash in one Jupyter Notebook
Combining Python, R, Bash in one Jupyter Notebook makes tracking of the workflow easier, simplifies sharing and makes you more efficient and professional.
Why Jupyter Notebooks?
If you read my Big Data tutorial, you are already familiar with Databricks notebooks. These notebooks allow combining code from many different programming languages (Scala, Python, etc.) in one notebook. I thought it would be great to set up a similar notebook environment locally on my computer to manage my workflows.
I used to log all my work steps in a text editor. It was a simple and reliable approach, but it was not the most efficient one. In particular, it was laborious to copy all commands I execute in the terminal to the text editor. Moreover, such log files were not user-friendly to share with my colleagues.
I knew that a Jupyter Notebook was the closest to Databriks notebook open-source solution that can be implemented on a desktop. But I knew Jupyter Notebooks only as Python Notebooks. However, after a few minutes searching online, I found out that one can easily combine Python, R, Bash in one Jupyter Notebook and execute code in all three languages within one notebook.
I would like to share with you how to set up such a notebook environment.
R and Bash in Jupyter cells
Below, I provide instruction for Linux OS, but I believe these commands are similar if not the same as in other operating systems.
First, install Jupyter Notebook. You can install it from your package manager by searching for the jupyter or by using pip:
python -m pip install jupyter
Second, install an R interface for Jupyter from your package manager by searching for rpy2 package or by using pip:
pip install rpy2
Third, load rpy2 in your Jupyter Notebook:
%load_ext rpy2.ipython
In my case, it complained that I had to install pandas as a dependency of rpy2. When I installed it, everything worked correctly.
Note: All these packages should be for the same Python version. So, keep track of whether you install these packages for Python 2 or Python 3.
After these simple steps, I was able to execute Python, R, Bash in one Jupyter Notebook by indicating R and Bash cells with %%R and %%bash, Jupyter magic commands.
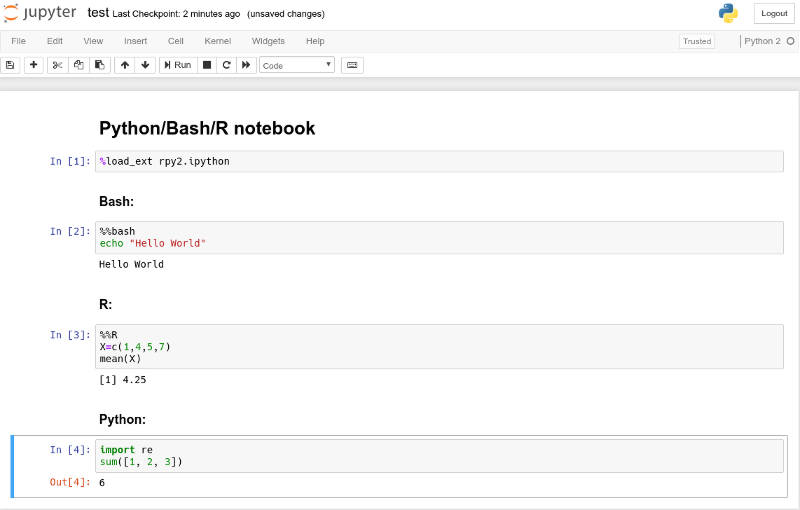
To test your installation, you can replicate my commands from the image above.
R and Bash Jupyter kernels
Basically, if you want to use Jupyter Notebooks primarily for Python code with an option to execute Bash and R, the steps described above are enough. However, you can also go further. If you install R and Bash Jupyter kernels, you will be able to use Jupyter Notebooks as notebooks for pipelines in either of these two languages.
To install a Jupyter kernel for Bash, execute:
pip install ipykernel
pip install bash_kernel
python -m bash_kernel.install
To install an R kernel for Jupyter, you need to run this code inside R:
install.packages('IRkernel')
IRkernel::installspec()
Now, you should be able to select a particular Jupyter kernel and create a Jupyter Notebook in either Python, Bash or R.
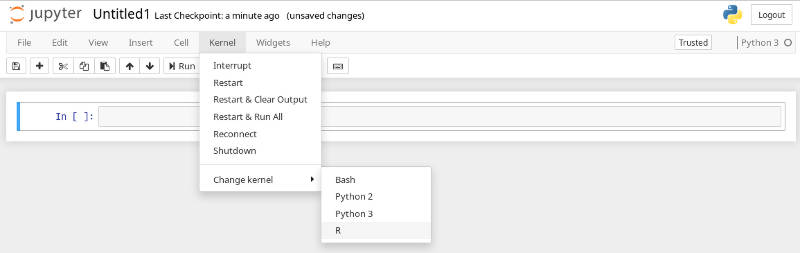
I usually use R and Bash kernels when I work on exclusive R or Bash pipelines.
Conclusion
This way you can use Jupyter Notebooks to log and execute your Python, R, Bash together in one single notebook as well as to create a well-annotated dedicated Python, R, Bash pipelines.
If you have any questions or suggestions, feel free to email me.