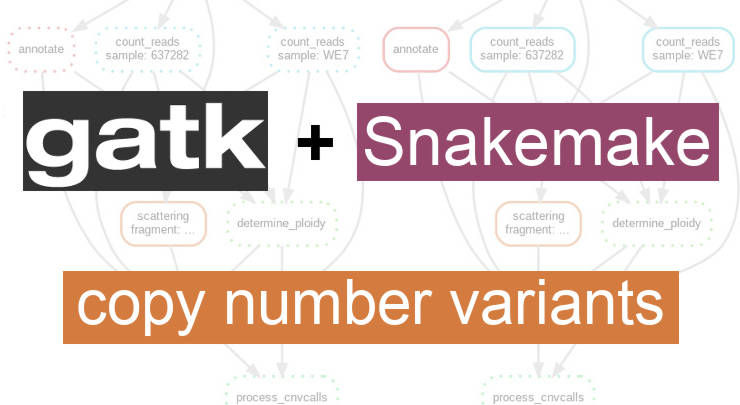
This pipeline calls germline copy number variants (CNV) with GATK 4 and Snakemake. It uses the cohort mode, so the CNV are inferred from all samples together....
Read More
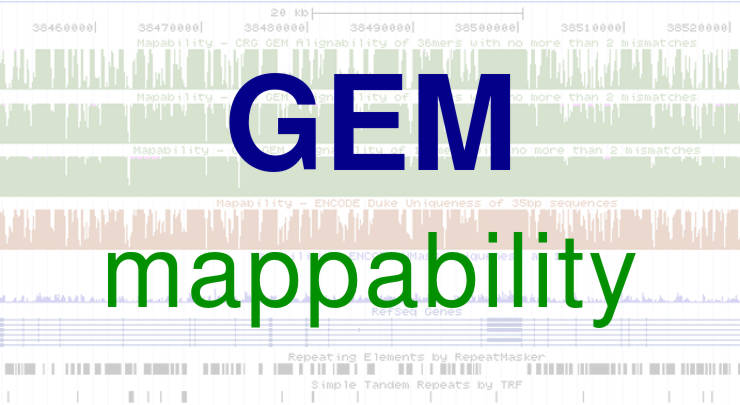
This post will guide you on how to get GEM mappability and convert it to BED file. Links to GEM library and all necessary conversion scripts are provided....
Read More
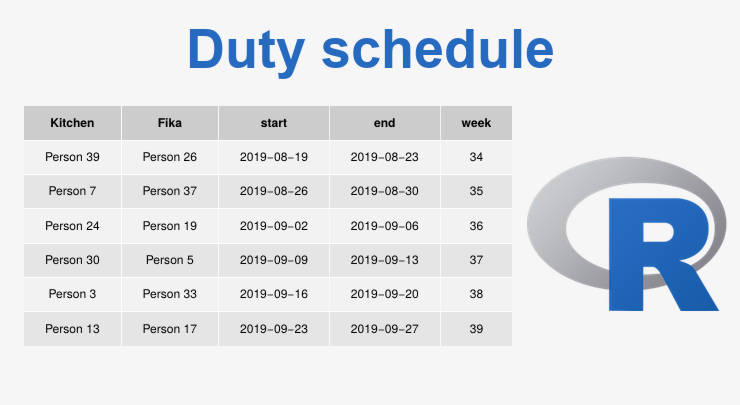
By creating a duty schedule in R you avoid bias and save time. You just provide a list of people and get a PDF of the duty schedule....
Read More

You can easily change the Docker default storage location by creating the daemon.json file and pointing to another location in that file....
Read More
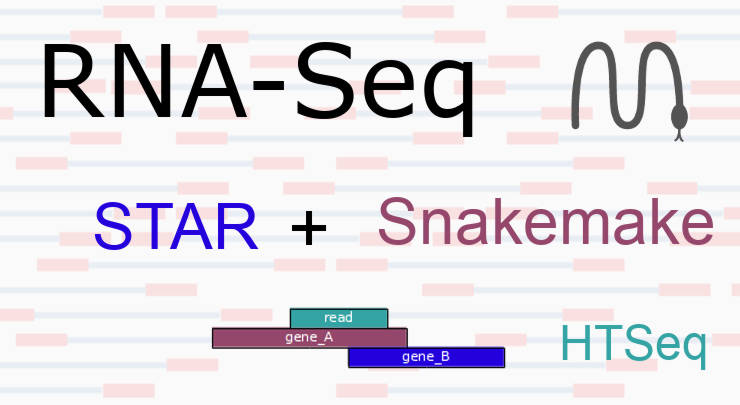
STAR mapping with Snakemake can save you a lot of time. STAR is a fast RNA-Seq aligner, whereas Snakemake provides automatic, reproducible, and scalable pipelining....
Read More




